The Idea of a Transform
Utilizing basic functions inside the DataBlock
This blog is also a Jupyter notebook available to run from the top down. There will be code snippets that you can then run in any environment. In this section I will be posting what version of fastai2 and fastcore I am currently running at the time of writing this:
fastai2: 0.0.13fastcore: 0.1.15
The DataBlock API Continued
This is part two of my series exploring the DataBlock API. If you have not read part one, fastai and the New DataBlock API, see here In it, we discussed the ideas of the DataBlock as a whole and how each of the lego-bricks can fit together to help solve some interesting problems. In this next blog, we’ll be slowly diving into more complex ideas and uses with it, such as adjusting our y values inside of our get_y, dealing with classification data seperated by folders (and the splitters we can use)
Also, as a little note, this blog is not explaining the Transform class, this will come later
From here on we’ll be focusing solely on generating the DataLoaders. Seperate blogs will be made about training the various models. Now, onto the code! As we’re still Vision based, we’ll use the vision sub-library:
from fastai2.vision.all import *ImageWoof
ImageWoof is a subset of 10 dogs from ImageNet. The idea is that these 10 species of dogs are extremely similar, and so they’re hard to classify from scratch. We won’t care about that part today, let’s go through and see how the data is formatted and apply the DataBlock. First let’s grab the data:
path = untar_data(URLs.IMAGEWOOF)Now if we take a look at the path first, we’ll notice that we have train and val folders. The two ideas I’ll be introducing with this dataset for splitting and labelling are GrandparentSplitter and parent_label
path.ls()What do each of these do? I’ll go into heavy detail on fastai’s splitters and labellers but GrandparentSplitter operates with the assumption our data is split like ImageNet, where we have training data in a training folder and validation data into a validation folder such as here. Let’s make a splitter now by passing in the name of the training folder and the validation folder:
splitter = GrandparentSplitter(train_name='train', valid_name='val')Let’s look at some splits. First we’ll grab our list of images then use our GrandparentSplitter to seperate out two indicies for us, which we’ll then look at to make sure it’s working properly
items = get_image_files(path)splits = splitter(items)splits[0][0], splits[1][0]Now let’s look at images 0 and 9025:
items[0], items[9025]And we can see that the folders line up!
Now that we have the splitter out of the way, we need a way to get our classes! But what do they look like? We’ll look inside the train folder at some of the images for some examples:
train_p = path/'train'train_p.ls()[:3]items = get_image_files(train_p)[:5]; itemsWe can visualize this folder setup like so:
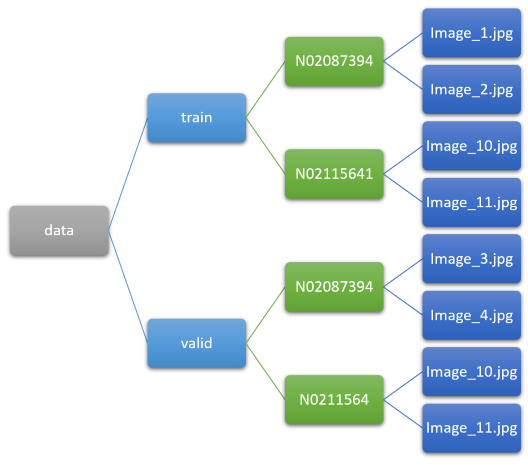
What this tells us is that our labels are in the folder one level above the actual image, or in the parent folder (if we consider it like a tree). As such, we can use the parent_label function to extract it! Let’s look:
labeller = parent_labellabeller(items[0])From here we can simply build our DataBlock similar to the last post:
blocks = (ImageBlock, CategoryBlock)
item_tfms=[Resize(224)]
batch_tfms=[Normalize.from_stats(*imagenet_stats)]block = DataBlock(blocks=blocks,
get_items=get_image_files,
get_y=parent_label,
item_tfms=item_tfms,
batch_tfms=batch_tfms)And make our DataLoaders:
dls = block.dataloaders(path, bs=64)To make sure it all worked out, let’s look at a batch:
dls.show_batch(max_n=3)The idea of a transform
Now we’re still going to use the ImageWoof dataset here, but I want to introduce you to the concept of a transform. From an outside perspective and what we’ve seen so far, this is normally limited to what we would call “augmentation.” With the new fastai this is no longer the case. Instead, let’s think of a transform as “any modification we can apply to our data at any point in time.”
But what does that really mean? What is a transform? A function! Any transform can be written out as a simple function that we pass in at any moment.
What do I mean by this though? Let’s take a look at those labels again. If we notice, we see bits like:
labeller(items[0]), labeller(items[1200])But that has no actual meaning to us (or anyone else reading to what we are doing). Let’s use a transform that will change this into something readable.
First we’ll build a dictionary that keeps track of what each original class name means:
lbl_dict = dict(n02086240= 'Shih-Tzu',
n02087394= 'Rhodesian ridgeback',
n02088364= 'Beagle',
n02089973= 'English foxhound',
n02093754= 'Australian terrier',
n02096294= 'Border terrier',
n02099601= 'Golden retriever',
n02105641= 'Old English sheepdog',
n02111889= 'Samoyed',
n02115641= 'Dingo'
)Now to use this as a function, we need a way to look into the dictionary with any raw input and return back our string. This can be done via the __getitem__ function:
lbl_dict.__getitem__(labeller(items[0]))But what is __getitem__? It’s a generic python function in classes that will look up objects via a key. In our case, our object is a dictionary and so we can pass in a key value to use (such as n02105641) and it will know to return back “Old English sheepdog” when called
Looks readable enough now, right? So where do I put this into the API. We can stack these mini-transforms anywhere we’d like them applied. For instance here, we want it done on our get_y, but after parent_label has been applied. Let’s do that:
block = DataBlock(blocks=blocks,
get_items=get_image_files,
get_y=[parent_label, lbl_dict.__getitem__],
item_tfms=item_tfms,
batch_tfms=batch_tfms)dls = block.dataloaders(path, bs=64)dls.show_batch(max_n=3)Awesome! It worked, and that was super simple. Does the order matter here though? Let’s try reversing it:
block = DataBlock(blocks=blocks,
get_items=get_image_files,
get_y=[lbl_dict.__getitem__, parent_label],
item_tfms=item_tfms,
batch_tfms=batch_tfms)dls = block.dataloaders(path, bs=64)
Oh no, I got an error! What is it telling me? That I was passing in the full image path to the dictionary before we extracted the parent_label, so order does matter in how you place these functions! Further, these functions can go in any of the building blocks for the DataBlock except during data augmentation (as these require special modifications we’ll look at later).